Suppose That We Have a Function F a R ‚ What Does It Mean for F to Be Continuous at C a
Useful R functions you might not know
Almost every R user knows about popular packages like dplyr and ggplot2. But with 10,000+ packages on CRAN and yet more on GitHub, it's not always easy to unearth libraries with great R functions. One of the best way to find cool, new-to-you R code is to see what other useRs have discovered. So, I'm sharing a few of my discoveries -- and hope you'll share some of yours in return (contact info below).
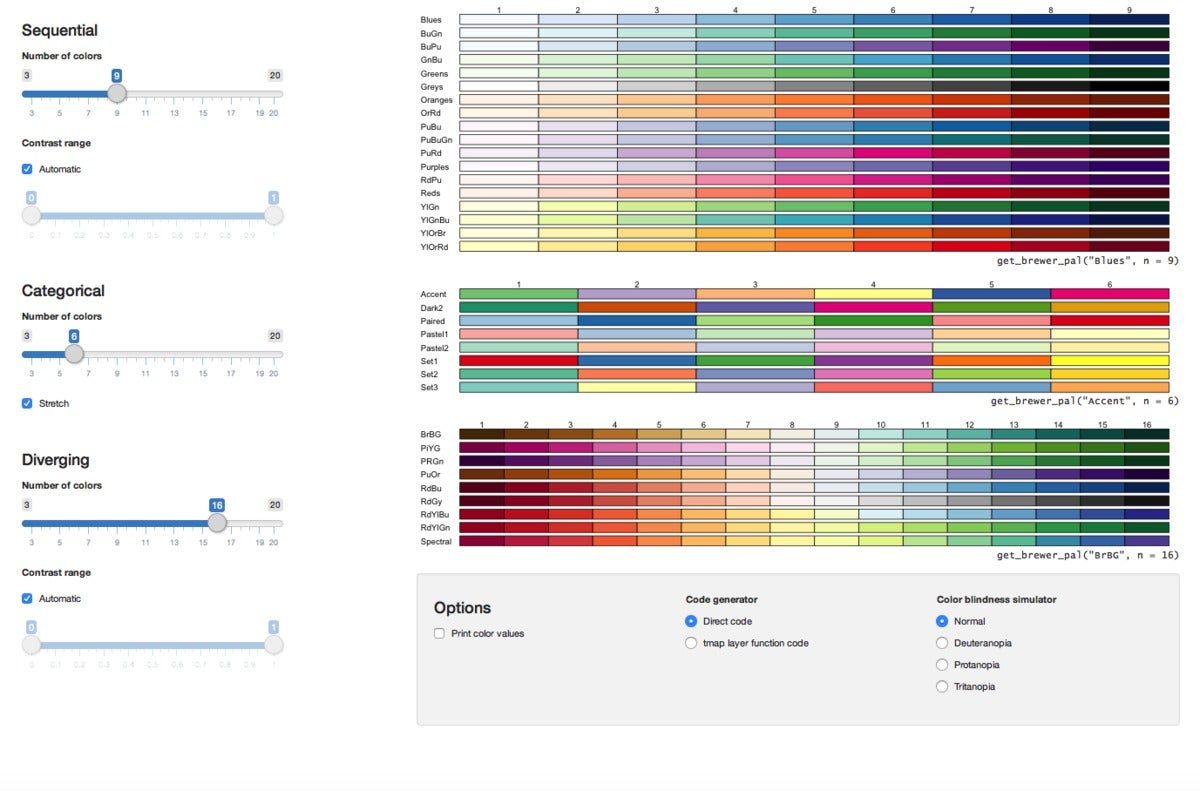
Choose a ColorBrewer palette from an interactive app. Need a color scheme for a map or app? ColorBrewer is well known as a source for pre-configured palettes, and the RColorBrewer package imports those into R. But it's not always easy to remember what's available. The tmaptools package's palette_explorer creates an interactive application that shows you the possibilities.
First, install tmaptools with install.packages("tmaptools"), then load tmaptools with library("tmaptools") and run palette_explorer() (or, don't load tmaptools and run tmaptools::palette_explorer() ). You'll see all available palettes as in the image above, as well as sliders to adjust options like number of colors. There's also info about basic syntax for using a color scheme below each group of palettes.
palette_explorer also needs shiny and shinyjs packages installed in order to generate the interactive app.
Create character vectors without quotation marks. It can be a bit annoying to manually turn Firefox, Chrome, Edge, Safari, Internet Explorer, Opera into the c("Firefox", "Chrome", "Edge", "Safari", "InternetExplorer", "Opera") format R needs to use such text as a vector of character strings.
That's what the Hmisc package's Cs function was designed to do. After loading the Hmisc package,
Cs(Firefox, Chrome, Edge, Safari, InternetExplorer, Opera) will evaluate the same as
c("Firefox", "Chrome", "Edge", "Safari", "InternetExplorer", "Opera") If you've ever manually added quotation marks to a lengthy string of words, you'll appreciate the elegance. Note the lack of a space in Internet Explorer -- spaces will trip up the Cs function.
RStudio bonus: If you use RStudio, there's another option for sleek vector-string creation. Security pro Bob Rudis created an RStudio add-in that takes selected comma-separated text and adds the necessary quotes and c(). And it can handle spaces. Install it with devtools::install_github("hrbrmstr/hrbraddins") (which means you need the devtools package as well), and you'll see Bare Combine as an option in the RStudio Tools > Addins menu.
You can run it from that Addins menu, but selecting text and then leaving your coding window to go to the Tools > Addins menu to select Bare Combine doesn't necessarily feel less cumbersome than typing a few quotation marks. Much better to create a custom keyboard shortcut for the addin.
You can do that by going to Tools > Modify Keyboard Shortcuts. Scroll down until you see Bare Combine in the Addins section -- or search for Bare Combine in the filter box. Double click in the shortcut area and type the keystroke(s) you want to assign to the addin (I usedalt-shift-').
Now, any time you want to turn comma-separated plain text into an R vector of character strings, you can highlight the text and use your keyboard shortcuts.
By the way, RStudio add-ins are mostly just plain R. If you'd like having keyboard shortcuts for R tasks like this, it might be worth learning the syntax.
Finally, the datapasta package's vector_paste() offers another unconventional alternative. You can copy a string like Firefox, Chrome, Edge, Safari, Internet Explorer, Opera into your clipboard and then run vector_paste(). That's it, just vector_paste(), and it converts your clipboard contents into R code, such as c("Firefox", "Chrome", "Edge", "Safari", "Internet Explorer", "Opera"). This works if there are tabs between the words as well as commas, or if each word is on its own line.
If you'd rather include data in your command, you can use vector_paste() with a syntax such as vector_paste("Firefox, Chrome, Safari, Edge") to generate the code such as c("Firefox", "Chrome", "Safari", "Edge"). datapasta has some other neat function, including df_paste(), which will turn a table copied into your clipboard from the Web, Excel, or other source, into code to generate a data frame.
Produce an interactive table with one line of code. Regardless of how much you like and use the command line, sometimes it's still nice to look at a spreadsheet-like table of data to scan, sort and filter. RStudio provided a basic view like this; but for large data sets, I like RStudio's DT package, a wrapper for the DataTables JavaScript library. DT::datatable(mydf) creates an interactive HTML table; DT::datatable(mydf, filter = "top") adds a filter box above each row.
Easy file conversions. rio is one of my favorite R packages. Instead of remembering which functions to use for importing what types of files (read.csv? read.table? read_excel?), rio vastly simplifies the process with one import function for a couple of dozen file formats. As long as the file extension is a format that rio recognizes, it will appropriately import from files such as .csv, .json, .xlsx and .html (tables). Same for rio's export command if you'd like to save to a particular file format. But rio has a third major function: convert, which will import and export in a single step. Have a million-row Excel file you need to save as a CSV? An HTML table you'd like to save as JSON? Use a syntax like convert("myfile.xlsx", "myfile.csv"), where the first argument is your existing file and the second is your desired file with the desired extension, and your file will be created.
Copy and paste from R to your clipboard. rio bonus: You can copy between your clipboard and R with rio. Send some data from a small R variable to your clipboard with export(myRobject, "clipboard"). Importing to the clipboard should work as well, although I've had mixed success with that.
Import large files quickly - and save space. It recently took close to 30 seconds when reading in a large spreadsheet. That's doable once, but annoying when I needed to access it multiple times. For saving space as well as wait time, the fst package was an excellent choice because it offers compression as well as high performance. In my testing, write.fst(mydf, "myfile.fst", 100) -- maximum compression -- was extremely quick -- and the .fst file took about one-third the space of the original spreadsheet.
Turn a data frame of numbers into one of percents. If you've got a data frame with one column of categories and the rest numbers -- imagine, say, a data frame showing election results by candidate and precinct -- the janitor package's adorn_percentages() will calculate all the percentages for you. You can choose whether the denominator for each percent should be summed by "row", "col" or "all". And, the function automatically assumes the first row has category information and skips it, without you having to manually deal with a non-numeric column.
janitor has several other handy functions worth knowing. adorn_totals() adds a totals row and/or column to a data frame. get_dupes() will find duplicated rows in a data frame based on one or more columns. And, clean_names() takes column names with spaces and other non-R-friendly characters in them and makes them R-compatible.
table() alternatives. Need to calculate frequencies of variables in a data frame? I like janitor's tabyl() function, which easily creates crosstabs with counts and percents and returns a data frame.
In addition, janitor's tabyl() can be used instead of base R's table(), helpfully returning a conventional data frame with counts and percents.
A few additional favorite functions from readers and social media:
"I'm a huge fan of xtabs()" for crosstabs, Timothy Teravainen posted at Google+. "It's in base R, but I sadly went years without knowing about it."
The format is xtabs(~df$col1 + df$col2), which will return a frequency table with col1 as the rows and col2 as the columns.
More with quotes. In response to the Cs() function that adds quotes, Kwan Lowe touted the usefulness of noquote(), which strips quotes -- useful for importing certain types of data into R. noquote() is a base R function, aimed it making it easier to wrangle variables.
Un-factoring factors. Another useful function: unfactor() in the varhandle package, which aims to detect the "real" class of an R data frame column of factors and then turn it into either numeric or character variables.
Text searching. If you've been using regular expressions to search for text that starts or ends with a certain character string, there's an easier way. "startsWith() and endsWith() -- did I really not know these?" tweeted data scientist Jonathan Carroll. "That's it, I'm sitting down and reading through dox for every #rstats function."
Loading packages -- and auto-installing if they're not present. For reproducible research, an R script can't simply load external packages -- it's got to check whether those packages are loaded on the user's machine and install them if they're not. There are several ways to do this in base R, such as using require() to check if various packages load and then installing the packages if they're not. The pacman package simplifies this immensely. To load packages and install them from CRAN if not available, the syntax is: p_load("package1", "package2", "package3"). There's also a p_load_gh() version for packages on GitHub. Thanks to Twitter user @Himmie_He for the tip.
Identifying your project's home directory. The here package's here() function finds the working directory for a current R project. This is especially handy for RStudio projects when a) your code needs access other directories and b) you'd like that code to work on other systems with a different directory structure. Thanks to Jenny Bryan and Hadley Wickham for that info via Twitter.
Get minimum and maximum values with a single command. Need to find the minimum and maximum values in a vector? Base R's range() function does just that, returning a 2-value vector with lowest and highest values. The help file says range() works on numeric and character values, but I've also had success using it with date objects.
Extract or operate on items in a list that are several layers deep. This is particularly useful if you're working with XML or JSON data imported into R, or you'd like to operate on multiple data frames but keep them separate. For example, this task tweeted by @netzstreuner asking if there was a better way to add a column to each data frame in a list of identically structured data frames:
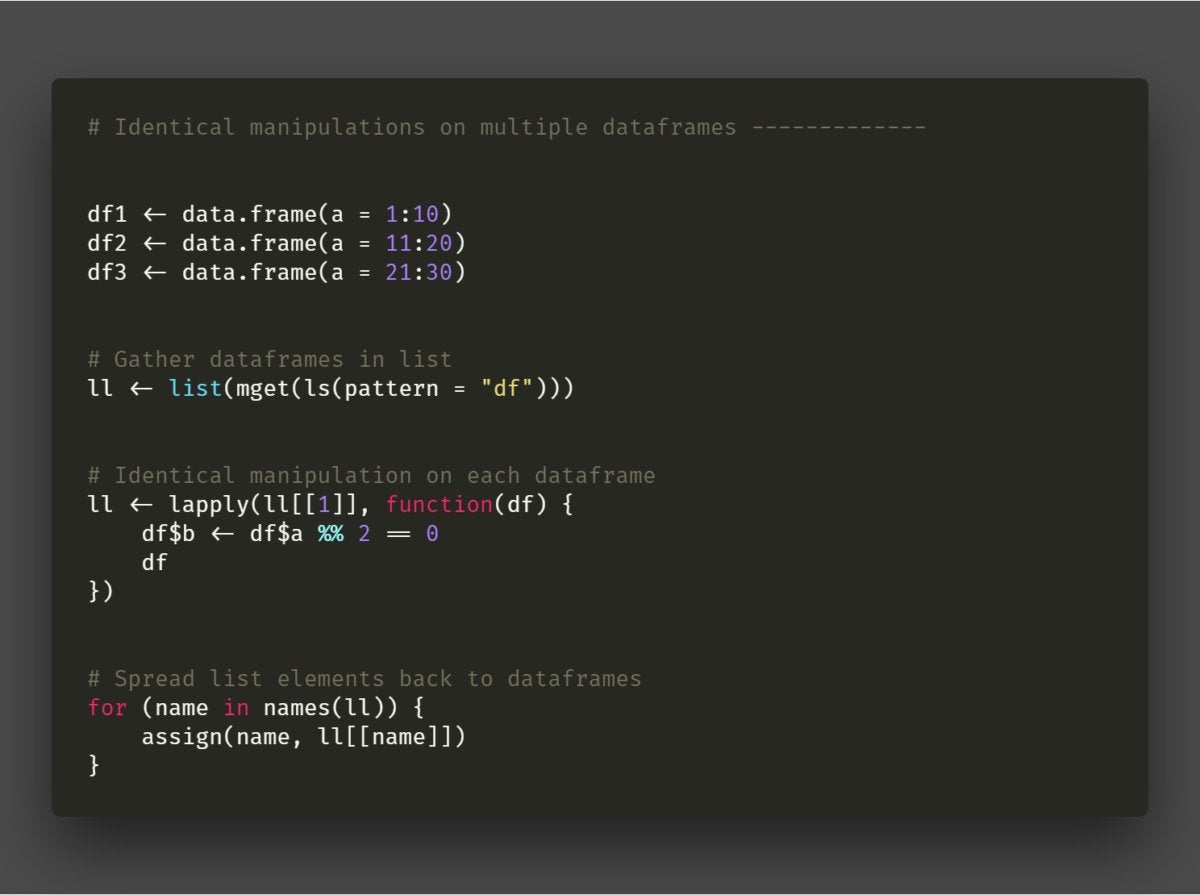 From @netzstreuner on Twitter
From @netzstreuner on Twitter Question from @netzstreuner on Twitter about operating on a specific column in every data frame within a list
The answer: purrr's modify_depth() function. modify_depth(mylist, 2, ~ myfunction) will run myfunction() on every item in mylist at the second level of that list.
That's for a generic list. Specifically for this question involving a list of data frames, dplyr's mutate() can add a new column to one data frame. To do this for a list of data frames, you can combine mutate() and modify_depth(). Here's my proposed solution to @netzstreuner's question:
ll_edited <- modify_depth(ll, 2, ~ mutate(.x, b = a %% 2 == 0)) That code says: "For every item two levels deep in the list ll, add a column b calculating if the value in column a is divisible by 2 with no remainder."
Easily filter a list. dplyr::filter() is a super-easy way to filter data frames. Have you ever wanted something similar for lists? Check out the rlist package's list.filter() function, which uses the syntax list.filter(mydf, mycondition) such as the package's example of list.filter(x, type == "B").
Get a number from a string. Have character strings that should be numbers? readr's parse_number() can handle formats such asparse_number("#3") andparse_number("1,012"). Columbia University stats lecturer Joyce Robbins noted on Twitter that you just want to be careful about negative numbers with certain formats. readr includes other handy parse_ functions, such as parse_time("4:30 pm").
Preview an R Markdown document each time you save. "Just a friendly reminder that xaringan:::inf_mr() works on any Rmd, and allows you to **live** preview your RMarkdown in the Viewer," data scientist Colin Fay tweeted. And that is indeed the case. Each time you save, a document will be re-generated automatically without specifically needing to knit or render.
Check user input when writing a function. Base R's match.arg() lets you input a vector of approved values for an argument, so users know that they've entered something that won't work instead of getting a more generic error message. That tip comes from Irene Steves' FUNctional programming tricks in httr tweeted by @dataandme.
Want to share your own favorites? Tell me via Twitter @sharon000 or email at sharon_machlis@idg.com.
For more on useful R functions, see Great R packages for data import, wrangling and visualization.
Copyright © 2018 IDG Communications, Inc.
Source: https://www.computerworld.com/article/3184778/6-useful-r-functions-you-might-not-know.html
0 Response to "Suppose That We Have a Function F a R ‚ What Does It Mean for F to Be Continuous at C a"
Post a Comment